低体温療法続き。設定温度からの解離が旧システムと新システムで異なるか検定しようと考えたが、正しい手法を選択できているか怪しい。検定以前に背景の患者因子が揃っていないことに対する注意を払っていないのが大問題だが、そもそもデータが12人分しか手に入らなかった... boxplotで箱ひげ図を描いた後に普通のplot関数を使って生データを○で描きたしている。箱ひげ図にも色を付けている。zorderを指定するとどの要素が上書きされるか指定できる。
追記(2018/7/1): L1 Errorの計算の際に記録体温がNAになってしまっている時間の除外を忘れてしまっていることに気付く
こちらについてもIDや時刻は匿名化済み。

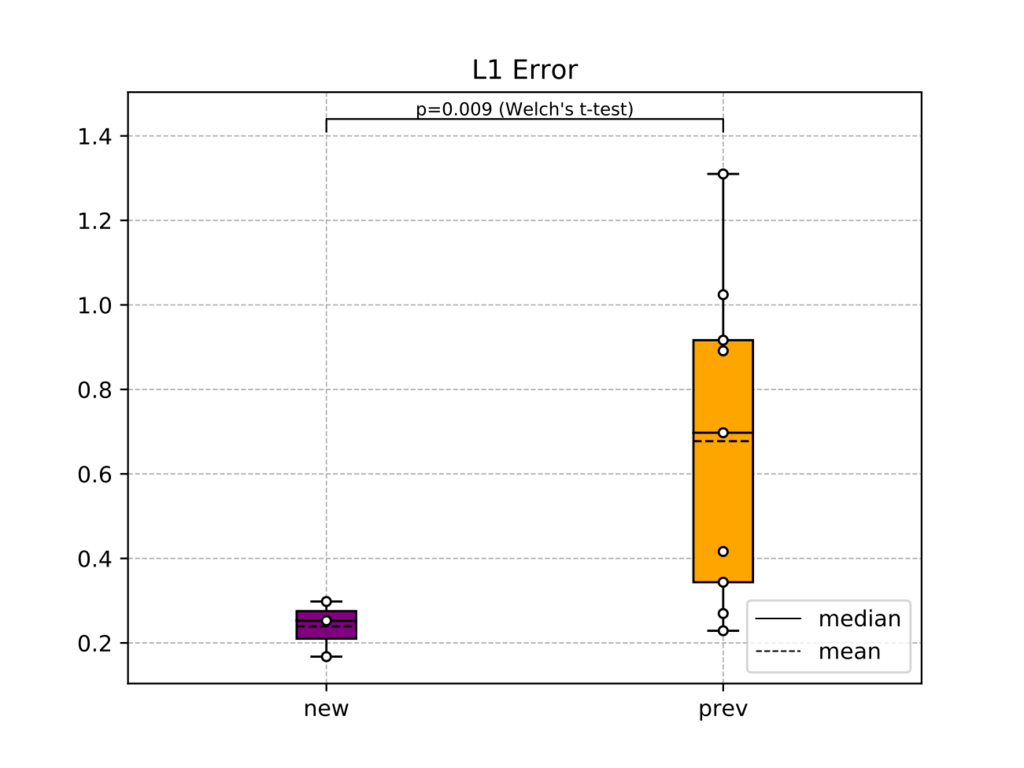
from pathlib import Path import pandas as pd import numpy as np import matplotlib.pyplot as plt import matplotlib.patches as mpatches plt.style.use('seaborn-muted') from scipy import stats ##################### # 旧システム ##################### filenames = [j for i in Path("./hypothermia/20XXMMDD_バイタル情報").iterdir() for j in i.iterdir()] patients_id_old_f = [str(i).split('/')[-1][6:].split('.')[0] for i in filenames] # [開始時間、復温開始時間、終了時間] record = { "1111111111": [pd.to_datetime(i) for i in ["20XX/MM/DD hh:mm", "20XX/MM/DD hh:mm", "20XX/MM/DD hh:mm"]],....} patients_id_old = list(record.keys()) def parse_time(s: str): s = s.replace("\n", "").split(',') date = "" ret = [] for i in s[1:-1]: if '/' in i: date = i.split(" ")[0] ret.append(i) else: ret.append(date + " " + i) return ret def parse_therm(s: str): s = s.replace("\n", "").split(',') ret = [] # 異常値の処理 for i in s[1:-1]: if i == "": ret.append(np.nan) elif 25 < float(i) < 45: ret.append(float(i)) else: ret.append(np.nan) return ret data_old = {} for fname, pid in zip(filenames, patients_id_old_f): with open(str(fname)) as f: is_time = False is_therm = False for i in f.readlines(): if i[:2] == "時刻": time = pd.to_datetime(parse_time(i)) is_time = True elif i[:2] == "T1": if sum([0 if np.isnan(i) else i for i in parse_therm(i)]): therm = parse_therm(i) is_therm = True if is_time and is_therm: data_old[pid] = [time, np.array(therm)] # 開始から6時間 data0_old = {} # 6時間後から復温開始まで data1_old = {} # 復温開始から終了まで data2_old = {} # 期間ごとに分類 for pid in patients_id_old: phase0_id = np.where((data_old[pid][0] > record[pid][0]) & (data_old[pid][0] <= record[pid][0] + np.timedelta64(6, 'h')))[0] phase1_id = np.where((data_old[pid][0] > record[pid][0] + np.timedelta64(6, 'h')) & (data_old[pid][0] <= record[pid][1]))[0] phase2_id = np.where((data_old[pid][0] > record[pid][1]) & (data_old[pid][0] <= record[pid][2]))[0] data0_old[pid] = [data_old[pid][0][phase0_id], data_old[pid][1][phase0_id]] data0_old[pid][0] = np.array([(i - record[pid][0])/np.timedelta64(1, 'D') for i in data0_old[pid][0]]) data1_old[pid] = [data_old[pid][0][phase1_id], data_old[pid][1][phase1_id]] data1_old[pid][0] = np.array([(i - record[pid][0] - np.timedelta64(6,'h'))/np.timedelta64(1, 'D') for i in data1_old[pid][0]]) data2_old[pid] = [data_old[pid][0][phase2_id], data_old[pid][1][phase2_id]] data2_old[pid][0] = np.array([(i - record[pid][1])/np.timedelta64(1, 'D') for i in data2_old[pid][0]]) # 維持期間の目標体温との差 for pid in patients_id_old: data1_old[pid][1] = np.abs(data1_old[pid][1] - 34.7) ##################### # 新システム ##################### filenames = [i for i in Path("./hypothermia/バイタルn人分/").iterdir()] patients_id_new = ["1111111",....] start = { "1111111": [pd.to_datetime("20XX/MM/DD hh:mm"), 34.5],....} change = { "1111111": [[pd.to_datetime("20XX/MM/DD hh:mm"), 34.8]]} start_off = { "1111111": [pd.to_datetime("20XX/MM/DD hh:mm"), 36.5],....} data_new = dict() for p in patients_id_new: data_new[p] = [] for f in filenames: if p in str(f): # データ読み込み df = pd.read_csv(str(f), index_col=0) data_new[p].append(df.loc['T1']) for p in patients_id_new: # 複数ファイル分のデータを連結する data_new[p] = pd.concat(data_new[p]) data_new[p] = data_new[p][~data_new[p].index.str.contains('^Unnamed') & ~data_new[p].index.duplicated()] data_new[p].index = pd.to_datetime(data_new[p].index) # 一応NaNの数を数えておく # print(p, data_new[p].isnull().sum()) # 異常値を削除してデータを日付順に並べ替える data_new[p] = data_new[p][(25 < data_new[p]) & (data_new[p] < 45)].sort_index(ascending=True) # 導入から6時間 data0_new = {} # 6時間後から復温開始まで data1_new = {} # 復温開始から終了まで data2_new = {} # 期間ごとに分類 for pid in patients_id_new: phase0_id = np.where((data_new[pid].index > start[pid][0]) & (data_new[pid].index <= start[pid][0] + np.timedelta64(6, 'h')))[0] phase1_id = np.where((data_new[pid].index > start[pid][0] + np.timedelta64(6, 'h')) & (data_new[pid].index <= start_off[pid][0]))[0] phase2_id = np.where(data_new[pid].index > start_off[pid][0])[0] data0_new[pid] = [data_new[pid].index[phase0_id], data_new[pid][phase0_id].values] data0_new[pid][0] = np.array([(i - start[pid][0])/np.timedelta64(1, 'D') for i in data0_new[pid][0]]) data1_new[pid] = [data_new[pid].index[phase1_id], data_new[pid][phase1_id].values] data1_new[pid][0] = np.array([(i - start[pid][0] - np.timedelta64(6,'h'))/np.timedelta64(1, 'D') for i in data1_new[pid][0]]) data2_new[pid] = [data_new[pid].index[phase2_id], data_new[pid][phase2_id].values] data2_new[pid][0] = np.array([(i - start_off[pid][0])/np.timedelta64(1, 'D') for i in data2_new[pid][0]]) #################### # データ分析 #################### # 維持期間の目標体温との差 for pid in patients_id_new: tmp = data1_new[pid][1] tmp = tmp - start[pid][1] if pid in change.keys(): for c in change[pid]: ct = (c[0] - start[pid][0])/np.timedelta64(1, 'D') tmp[data1_new[pid][0] > ct] = data1_new[pid][1][data1_new[pid][0] > ct] - c[1] data1_new[pid][1] = np.abs(tmp) plt.title("L1 deviation from set tempareture") for pid in patients_id_old: plt.plot(data1_old[pid][0], data1_old[pid][1], color="orange") for pid in patients_id_new: plt.plot(data1_new[pid][0], data1_new[pid][1], color="purple") plt.xlabel('day', position=(1.05,0)) plt.ylabel('℃', position=(0, 1), rotation=0) plt.savefig("l1_graph.svg") plt.clf() plt.title("L2 deviation from set tempareture") for pid in patients_id_old: plt.plot(data1_old[pid][0], data1_old[pid][1]**2, color="orange") for pid in patients_id_new: plt.plot(data1_new[pid][0], data1_new[pid][1]**2, color="purple") plt.xlabel('day', position=(1.05,0)) plt.ylabel('℃', position=(0, 1), rotation=0) plt.savefig("l2_graph.svg") plt.clf() def diff(xs, ys): if len(xs) != len(ys): raise ValueError() ret = [xs[:-1], np.empty(len(xs)-1)] for i in range(len(xs)-1): ret[1][i] = (xs[i+1] - xs[i]) / (ys[i+1] - ys[i]) return ret def integrate(xs, ys): if len(xs) != len(ys): raise ValueError() ret = 0 x_tmp, y_tmp = xs[0], ys[0] xmin = 0 for i in range(len(xs)-1): if not np.isnan(ys[i]): xmin = i break for i in range(xmin, len(xs)-1): if not np.isnan(ys[i+1]): ret += (xs[i+1] - x_tmp) * (ys[i+1] + y_tmp) / 2 x_tmp, y_tmp = xs[i+1], ys[i+1] return ret l1_error_new = [integrate(*data1_new[pid])/(data1_new[pid][0][-1] - data1_new[pid][0][0]) for pid in patients_id_new] l1_error_old = [integrate(*data1_old[pid])/(data1_old[pid][0][-1] - data1_old[pid][0][0]) for pid in patients_id_old] l2_error_new = [integrate(data1_new[pid][0], data1_new[pid][1]**2) for pid in patients_id_new] l2_error_old = [integrate(data1_old[pid][0], data1_old[pid][1]**2) for pid in patients_id_old] max_l1 = np.max(l1_error_new + l1_error_old) max_l2 = np.max(l2_error_new + l2_error_old) #equal_var=Trueで分散が等しい場合のt検定、FalseでWelchのt検定 t1, p1 = stats.ttest_ind(l1_error_new, l1_error_old, equal_var=False) t2, p2 = stats.ttest_ind(l2_error_new, l2_error_old, equal_var=False) # print("t-value = " + str(t)) # print("p-value = " + str(p)) patch0 = plt.plot([], [], label="median", linestyle="-", color="k", linewidth=0.7)[0] patch1 = plt.plot([], [], label="mean", linestyle="--", color="k", linewidth=0.7)[0] meanprops = dict(color="k", linestyle="--") medianprops = dict(color="k", linestyle="-") plt.title("L1 Error") plt.grid(linestyle='--', linewidth=0.6) # 箱ひげ図 bp = plt.boxplot( [l1_error_new, l1_error_old], labels=["new", "prev"], meanprops=meanprops, meanline=True, showmeans=True, medianprops=medianprops, showfliers=False, patch_artist=True, zorder=2 ) for i, patch in enumerate(bp['boxes']): patch.set_facecolor(["purple", "orange"][i]) # 実データもプロット for i in (0, 1): data = [l1_error_new, l1_error_old] x = [i+1]*len(data[i]) y = data[i] plt.plot(x, y, 'k.', markerfacecolor='#ffffff', markersize=8, zorder=3) # p値の下の棒 plt.plot([1, 1, 2, 2], [max_l1+0.1, max_l1+0.13, max_l1+0.13, max_l1+0.1], linewidth=0.8, color="black") # p値を書き込む plt.text(1.5, max_l1+0.13, "p={0:.3f} (Welch's t-test)".format(p1), ha='center', va='bottom', color="black", fontsize=8) plt.legend(handles=[patch0, patch1], loc="lower right") plt.savefig("l1error_boxplot.svg") plt.clf() plt.title("L2 Error") plt.grid(linestyle='--', linewidth=0.6) # 箱ひげ図 bp = plt.boxplot( [l2_error_new, l2_error_old], labels=["new", "prev"], meanprops=meanprops, meanline=True, showmeans=True, medianprops=medianprops, showfliers=False, patch_artist=True, zorder=2 ) for i, patch in enumerate(bp['boxes']): patch.set_facecolor(["purple", "orange"][i]) # 実データもプロット for i in (0, 1): data = [l2_error_new, l2_error_old] x = [i+1]*len(data[i]) y = data[i] plt.plot(x, y, 'k.', markerfacecolor='#ffffff', markersize=8, zorder=3) # p値の下の棒 plt.plot([1, 1, 2, 2], [max_l2+0.1, max_l2+0.13, max_l2+0.13, max_l2+0.1], linewidth=0.8, color="black") # p値を書き込む plt.text(1.5, max_l2+0.13, "p={0:.3f} (Welch's t-test)".format(p1), ha='center', va='bottom', color="black", fontsize=8) plt.legend(handles=[patch0, patch1], loc="lower right") plt.savefig("l2error_boxplot.svg") plt.clf()